Site-Directed Mutagenesis on a Minicircle using Overlap Extension PCR
PCR Success Story #22Project Description
Generate double mutants in a minicircle vector with contains very few unique restriction sites.
(paid service)
The goal was to combine mutations in a minicircle vector coding for the genes of a Human Papilloma Virus Strain.
The researcher supplied us with 3 single mutant vectors, in which we needed to introduce a downstream mutation that would abolish the start codon for a second gene encoded within the first gene but in a different reading-frame.
Project Details
Client: McGill University
Date: April 1rst, 2017
Type of experiment: Create double mutants in a minicricle vector
DNA Polymerase: FastPfu FLY
Competitor: N/A
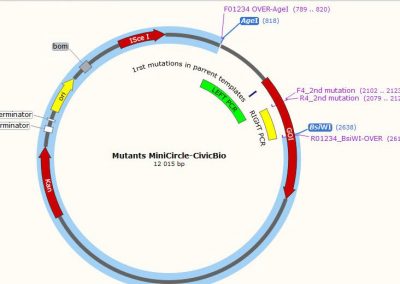
Vector Restriction Digest
Minicircle Vector Restriction Digest
Minicircle Vector Restriction Digest with BsiWI and AgeI. To maximize the chance of achieving full parental DNA digestion, we recommend performing a first digestion in 50 ul for 1h followed by DNA clean-up. Then, a second identical digestion for 15min, followed by agarose gel electrophoresis and gel extraction.
Full Restriction Digest of the Backbone Vector
1rst Digestion (60 min)
ddH2O : to 50 ul
10x buffer : 5 ul
Vector : 2 ug
AgeI-HF : 1.5 ul
BsiWI : 1.5 ul
DNA Clean-Up
Follow the instructions of FavorPrep GEL/PCR Purification Kit or FavorPrep MicroElute GEL/PCR Purification Kit and elute the DNA in 40 ul EB buffer.
2nd Digestion (15 min)
ddH2O : 3 ul
10x buffer : 5 ul
Eluate : 40 ul
AgeI-HF : 1 ul
BsiWI : 1 ul
DNA Clean-Up
- Mix 10 ul of 6x DNA loading buffer with your sample and load on a 0.7 % agarose gel prestained with GelStain.
- Cut out the band using a clean razor blade and follow the instructions for FavorPrep GEL/PCR Purification Kit. Elute the DNA in 40 ul EB buffer.
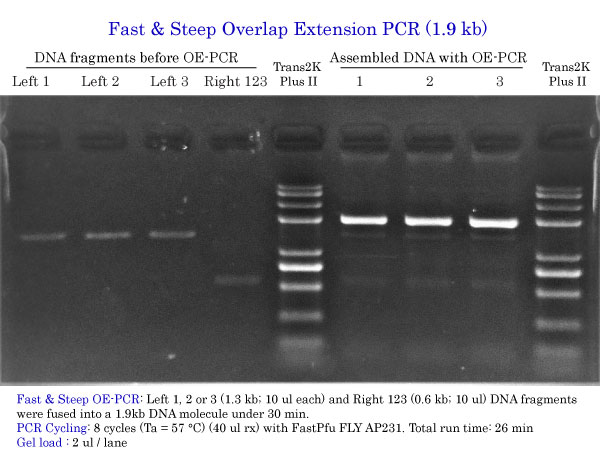
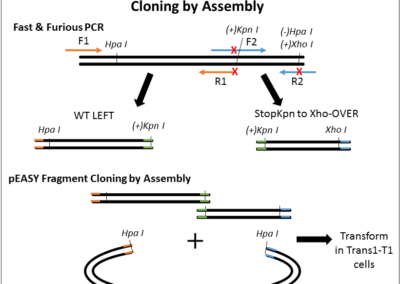
PCR used to Generate a Mutation and Create Overlapping DNA Fragments
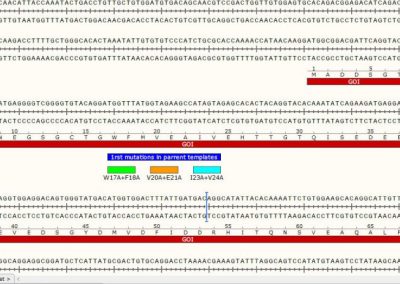
DNA sequence segment showing the location of 3 individual mutations present in 3 individual Minicircles.
Only 1 mutation is present in each three different parental plasmid templates. All three are depicted here on the same DNA sequence for simplicity.
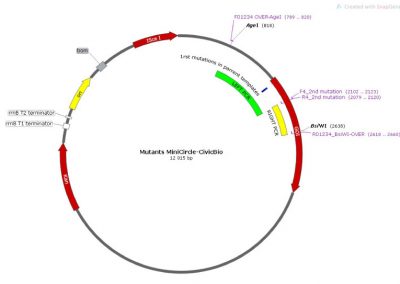
Map of the minicircle templates used for overlap extension PCR.
3 individual minicircles are used as template for PCR for combining the 1rst mutation (either 3 of them) with a 2nd mutation. The region amplified by the LEFT PCRs (for Left 1, 2 and 3) is shown in green. The common RIGHT PCR region used to add the 2nd mutation is shown in yellow. The 3 LEFT PCRs and the RIGHT PCR were amplified using Fast & Steep PCR.
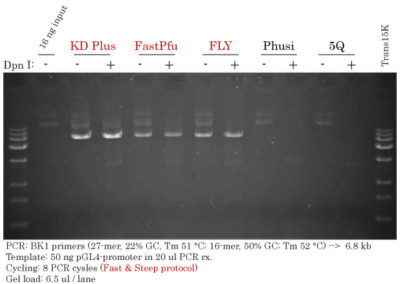
Fast & Steep PCR to generate LEFT and RIGHT fragments
PCR Setup :
ddH2O : to 35 ul
5x buffer: 7 ul
dNTPs (2,5 mM) : 2.8 ul
F primer (100 uM) : 0.14 ul (14 pmol)
R primer (100 uM) : 0.14 ul (14 pmol)
*Minicircle : 300 ng
FastPfu FLY : 0.7 ul (1.75 u)
PCR Cycling: 7 cycles (Ta = 62 °C)
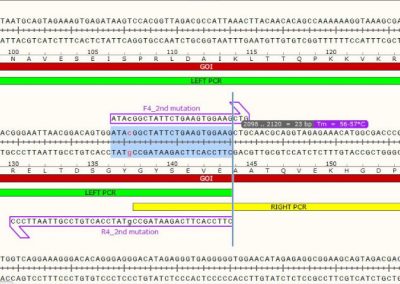
Overlap between Left and Right DNA fragments before OE-PCR
LEFT and RIGHT PCR fragments overlap each other by 23 bp. The overlapping region contains the second desired mutation in order to generate double mutants. The 23 bp overlap has an expected Tm of 57 °C. It’s important to design overlapping DNA segments having a Tm equal or greater than the Tm of external primers used for the OE-PCR reaction.
DNA Assembly using PCR Overlap Extension
Fast & Steep OE-PCR 1.9 kb
PCR Setup :
ddH2O : to 40 ul
5x buffer: 8 ul
dNTPs (2,5 mM) : 3.2 ul
F primer (10 uM) : 1.6 ul (16 pmol)
R primer (10 uM) : 2 ul (16 pmol)
LEFT DNA : 10 ul
RIGHT DNA : 10 ul
FastPfu FLY : 0.8 ul (2 u)
PCR Cycling: 8 cycles (Ta = 57 °C)
Total run time: 26 min
Gel load : 2 ul / lane
Cloning of the Overlapped DNA into the Linearized Vector
Seamless Assembly of a DNA Fragment and a Linearized Vector
2x assembly mix : 2.5 ul
Linearized vector : 0.4 ul
Insert : 1.1 ul
Incubation : 20 min at 50 °C
Transformation of the Fully Assembled Minicircles into Competent Cells
Plasmid Transformation into Trans2-Blue Chemically Competent Cells
- Thaw 50 ul of cells on ice.
- Add 1.5 ul of the pEASY-Uni reaction mix
- Gently flick the tube twice, then put on ice for 30 min
- Heat Shock for 30 s at 42 °C in a water bath.
- Put on ice for 2 min.
- Add 450 ul of room temperature SOC or LB media.
- Shake at 200 rpm at 37 °C for 45 min.
- Spread 200 ul on a pre-warmed kanamycin-selective LB-agar plate.
- Incubate O/N at 37 °C
* We typically spread 60-100 ul on LB-agar plates, but this particuliar minicircle vector has very poor transformation efficiency in all strains tested.
Clone Validation by Restriction Analysis and Sanger Sequencing
Plasmid Minipreps and Restriction Analysis
Plasmid Minipreps
Two to four colonies per plate were picked and inoculated in 2.5 ml LB media and selective for kanamycin-resistant bacteria.
An aliquot of 850 ul was reserved for the glycerol stock and the remaining bacteria (≤ 1.65 ml) served for plasmid DNA extraction using FAPDE-300 from Favorgen.
The DNA was eluted in 50 ul EB solution.
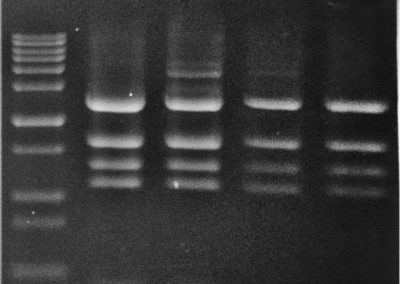
Restriction Analysis
ddH2O : to 10 ul
10x CS buffer : 1 ul
Plasmid DNA : 2 ul
Bsu36I : 0.3 ul
NdeI : 0.3 ul
Incubated 15 min at 37 °C.