Fast & Steep PCR Protocol
Fast PCR Protocol for Cloning and Site-Directed MutagenesisFast & Steep PCR can be used for quick DNA amplification from plasmid DNA, purified DNA fragments and assembly PCR by extension of overlapping DNA fragments.
DNA Amplification by PCR is Exponential

A typical PCR reaction is divided into 3 steps and the second step is divided also into 3 steps.
- Initial denaturation at 94-98 °C for 30s to 300s
- Repeated Amplification
- Denaturation at 94-98 °C for 5s to 30s
- Annealing at Ta °C for 5s to 30s
- Extension at 66-75 °C for 30/60s per kb
- Final Extension at 66-75 °C for 120s to 600s
The basis behind the Fast & Steep PCR protocol
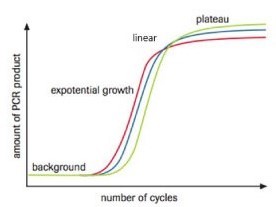
Fast & Steep PCR takes advantage of the exponential growth rate of DNA amplification by PCR.
The Fast & Steep PCR protocol will give you this
Standard PCR Protocol
Standard PCR reaction setup
ddH2O : to 50 ul
5x buffer: 10 ul
dNTPs (2,5 mM) : 4 ul
F primer (10 uM) : 1 ul (0,2 uM) )(10 pmol)
R primer (10 uM) : 1 ul (0,2 uM) )(10 pmol)
DNA (plasmid or fragment): 1 pg to 50 ng
FastPfu FLY : 1 ul (2.5 u)
Standard PCR Cycling
1-5 min denaturation at 95°C
25-35x:
15-30s denaturation at 95°C
10-30s annealing at optimal Ta
10-30s/kb (2-6 kb/min) at 72°C
2-10 min final extension at 72°C
Fast & Steep PCR Protocol
Fast & Steep PCR reaction setup
ddH2O : to 50 ul
5x buffer: 10 ul
dNTPs (2,5 mM) : 4 ul
F primer (10 uM) : 2 ul (0,4 uM)(20 pmol***)
R primer (10 uM) : 2 ul (0,4 uM)(20 pmol***)
DNA (plasmid or fragment): 100 ng to 1 ug (0,25 to 0,5 pmol***)
FastPfu FLY : 1 ul (2.5 u)
Fast & Steep PCR Cycling
2-5 min denaturation at 95°C
5-10x :
20s denaturation at 95°C
30s annealing at Ta of highest primer Tm
30-60s/kb (1-2 kb/min) at 68°C
5 min final extension at 68°C
Advantages of using the Fast & Steep PCR Protocol
Very Fast PCR Protocol
- 15-45 min for 300 bp and 10 kb respectively.
- PCR cycling time varies depending on template lenght and ramping rates.
Efficient DNA amplification
- Incorporates up to 100% of primers in a very short amount of time.
- GC content and primer specificity may affect the efficiency.
Non-Ambiguious PCR Results
- PCR amplicons have fully incorporated your primers.
- When performing site-directed mutagenesis, so far, our success rate is 100%.
Low error rate / High-Fidelity PCR
- 5 to 10 PCR cycles is sufficient.
- Limit the chance of introducing errors by limiting the amount of doublings and heating cycles, without compromising the DNA yield.
Using the fewest number of PCR cycles helps to avoid DNA depurination and deamination
Depurination
⇒ Depurination involves the loss of purine bases forming abasic sites.
⇒ Depurination decreases at higher pH.
⇒ Depurination is independent of DNA sequence.
⇒ Heating DNA for 10 minutes at 100°C with pH 7.0 leads to about 1 apurinic site per 1000 base pairs.
Deamination
⇒ Cytosine can be spontaneously deaminated to form uracil.
⇒ Cytosine in native DNA is estimated to deanimate with a rate constant of 10-10/sec at 70°C.
Reason #1
Our very first Fast & Steep PCR
Reason #2
Reason #3
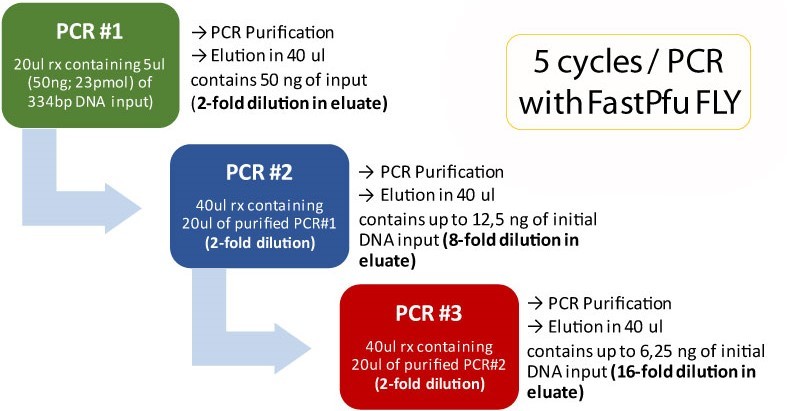
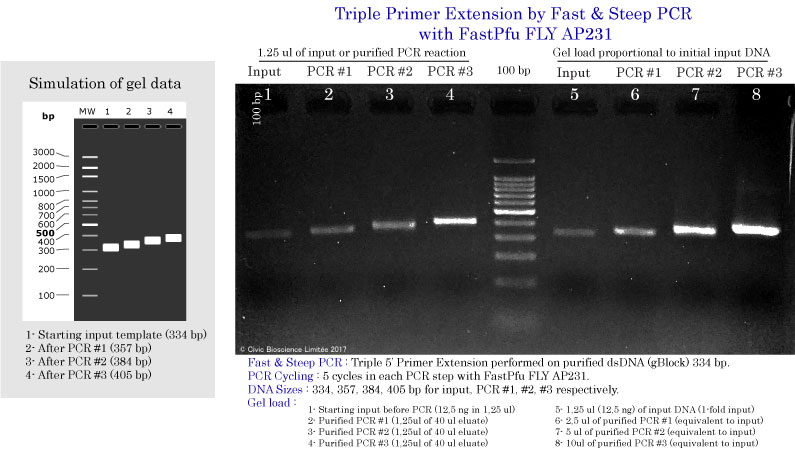
Triple primer extension using Fast & Steep PCR
DNA input and DNA Polymerases used in Fast & Steep PCR
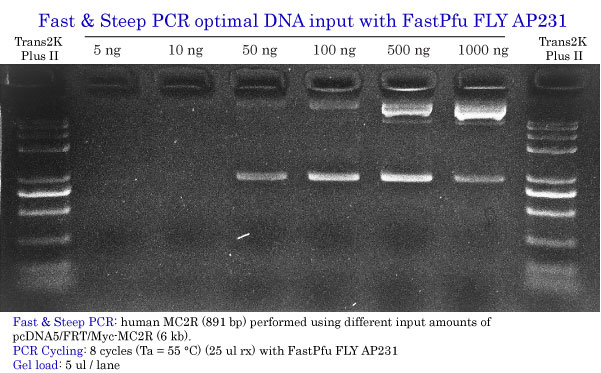
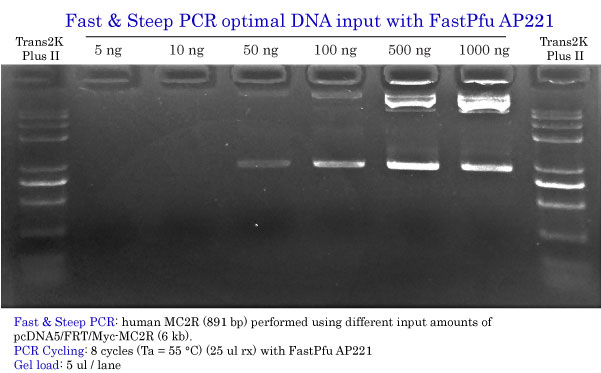
Determination of the Optimal DNA input
Fast & Steep PCR with FastPfu FLY
Fast & Steep PCR with FastPfu
Tip on Primer Design for Efficient DNA Amplification by PCR
Use a Dedicated Design Software
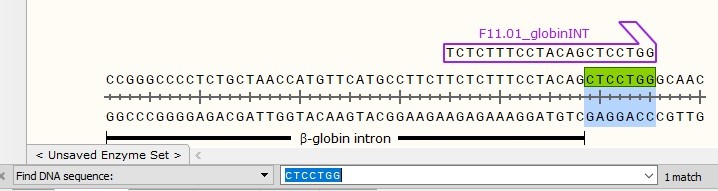
GC-clamp
Does a sprint runner start its race on its back foot? No! In order to start its race, a sprint runner puts most of his weight on his front foot in order to create strong friction with the ground and get a powerful start to the finish line.
DNA Polymerases also require to grip the DNA template tightly for initiating 5′ to 3′ polymerization. Using a GC-clamp at the 3′ end helps to create this tight grip on single-stranded DNA strand to initiate elongation.
A good GC-clamp consists in ending the 3′ end of the DNA primer with 1 or 2 ‘G’ or ‘C’ (in bold below).
i.e: TCTCTTTCCTACAGCTCCTGG
The 7-base rule
What can I Achieve by using Fast & Steep PCR ?
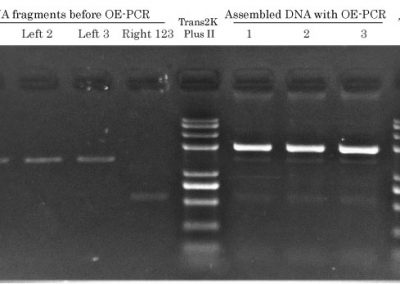
Assemble Overlapping DNA Fragments? Sure!