Overlap Extension PCR Protocol
PCR Protocol for DNA assembly by PCR extension of overlapping DNA fragments.- a standard protocol for performing overlap extension PCR
- our Fast & Steep PCR protocol for overlapping DNA fragments
Overlap Extension Polymerase Chain Reaction
Splicing of DNA Molecules
As in most PCR reactions, two primers are per target DNA sequence are required. To splice two DNA molecules, custom primers are used to extend DNA ends that are to be joined. For each molecule, the primer at the end to be joined is constructed such that it has a 5′ overhang complementary to the end of the other DNA fragment. Following annealing when replication occurs, the DNA is extended by a new sequence that is complementary to the DNA molecule. Once both DNA molecules are extended in such a manner, they are mixed and a PCR is carried out with only the primers for the far ends. The overlapping complementary sequences introduced will serve as primers and the two sequences will be fused. This method has an advantage over other gene splicing techniques in not requiring restriction sites. To get higher yields, some primers are used in excess as in asymmetric PCR. Ref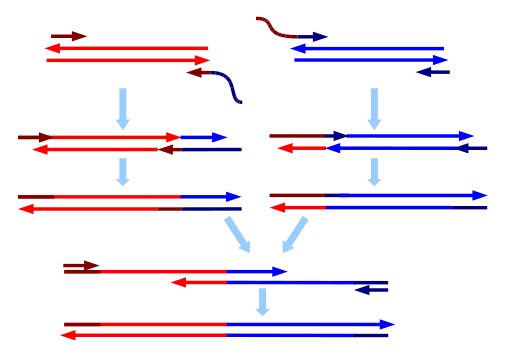
Overview of Overlap Extension PCR
Why perform Overlap Extension PCR?
At Civic Bioscience, we often use OE-PCR to fuse multiple DNA sequences together for the purpose of:- Cloning : a complete gene cannot be amplified from cDNA because specific sequence segments are too different in composition (i.e GC content) and are very difficult to amplify when the amount of target DNA is very low.
- Site-Directed Mutagenesis : performing DNA mutagenesis using a 2-sided PCR overlap extension followed by ligation or seamless assembly into a linerized vector.
- Multiple gene fusions, cassette or plasmid engineering : things can become quite complex when we engineer custom clones for our customers. Assembling 9 DNA fragments together by seamless assembly (i.e Gibson assembly) won’t work efficiently. But, if assembly by OE-PCR is used to put together fragments in groups of 3, then seamless DNA assembly using pEASY-Uni will become easy enough to get our clones rapidly. We use the Fast & Steep PCR protocol to accomplish this.
pEASY®-Uni Seamless Cloning and Assembly Kit
Expected Results from Overlap Extension PCR used to join 2 DNA fragments of different sizes
Standard Overlap Extension PCR Protocol
Standard PCR reaction setup
ddH2O : to 50 ul
5x buffer: 10 ul
dNTPs (2,5 mM) : 4 ul
F primer (10 uM) : 1 ul (0,2 uM) )(10 pmol)
R primer (10 uM) : 1 ul (0,2 uM) )(10 pmol)
Fragment 1 : 1 ul and equimolar to 2 and 3
Fragment 2 : 1 ul and equimolar to 1 and 3
Fragment 3 : 1 ul and equimolar to 1 and 2
FastPfu FLY : 1 ul (2.5 u)
Standard PCR Cycling
1-5 min denaturation at 95°C
25-35x:15-30s denaturation at 95°C
10-30s annealing at optimal Ta
10-30s/kb (2-6 kb/min) at 72°C
2-10 min final extension at 72°CFast & Steep OE-PCR Protocol
Fast & Steep PCR reaction setup
ddH2O : to 50 ul
5x buffer: 10 ul
dNTPs (2,5 mM) : 4 ul
F primer (10 uM) : 2 ul (0,4 uM)(20 pmol***)
R primer (10 uM) : 2 ul (0,4 uM)(20 pmol***)
Fragment 1 : 20-30% of final volume and equimolar to 2 and 3
Fragment 2 : 20-30% of final volume and equimolar to 1 and 3
Fragment 3 : 20-30% of final volume and equimolar to 1 and 2
FastPfu FLY : 1 ul (2.5 u)
Fast & Steep PCR Cycling
2-5 min denaturation at 95°C 5-10x :20s denaturation at 95°C
30s annealing at Ta of highest primer Tm
30s-60s/kb (2 kb/min) at 68°C
5 min final extension at 68°CWhy you should use the Fast & Steep PCR protocol for your next Overlap Extension PCR
Very Fast PCR Protocol
- 15-45 min for 300 bp and 10 kb respectively.
- PCR cycling time varies depending on template lenght and ramping rates.
Efficient DNA amplification
- Incorporates up to 100% of primers in a very small amount of PCR cycles.
- GC content and primer specificity may affect the efficiency.
Easy assembly of DNA fragments
- Get your assembled DNA in 30 min, on average
Low error rate / High-Fidelity PCR
- 5 to 10 PCR cycles is sufficient.
- Limit the chance of introducing errors by limiting the amount of doublings and heating cycles, without compromising the DNA yield.
Using the fewest number of PCR cycles helps to avoid DNA depurination and deamination
Depurination
⇒ Depurination involves the loss of purine bases forming abasic sites.
⇒ Depurination decreases at higher pH.
⇒ Depurination is independent of DNA sequence.
⇒ Heating DNA for 10 minutes at 100°C with pH 7.0 leads to about 1 apurinic site per 1000 base pairs.
Deamination
⇒ Cytosine can be spontaneously deaminated to form uracil.
⇒ Cytosine in native DNA is estimated to deanimate with a rate constant of 10-10/sec at 70°C.
Examples of DNA assembly using PCR overlap extension
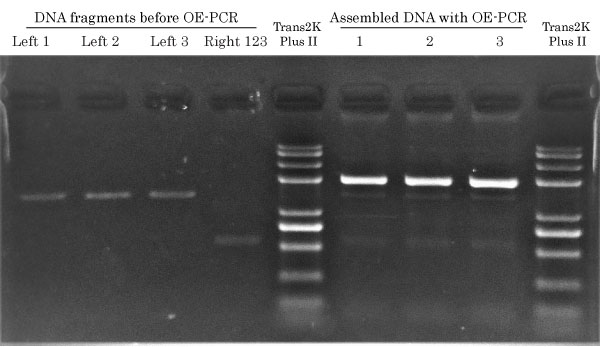
Results from PCR Success Story #22
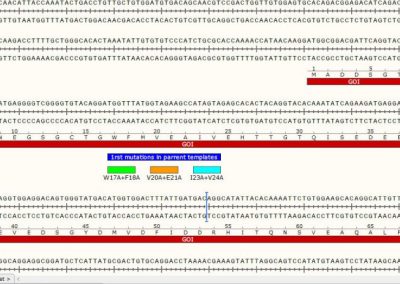
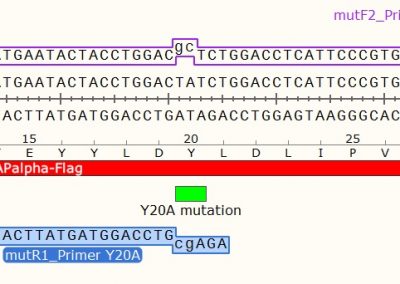
DNA sequence segment showing the location of 3 individual mutations present in 3 individual Minicircles.
Only 1 mutation is present in each three different parental plasmid templates. All three are depicted here on the same DNA sequence for simplicity.
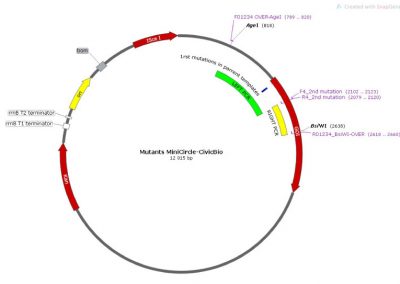
Map of the minicircle templates used for overlap extension PCR.
3 individual minicircles are used as template for PCR for combining the 1rst mutation (either 3 of them) with a 2nd mutation. The region amplified by the LEFT PCRs (for Left 1, 2 and 3) is shown in green. The common RIGHT PCR region used to add the 2nd mutation is shown in yellow. The 3 LEFT PCRs and the RIGHT PCR were amplified using Fast & Steep PCR.
Fast & Steep PCR to generate LEFT and RIGHT fragments
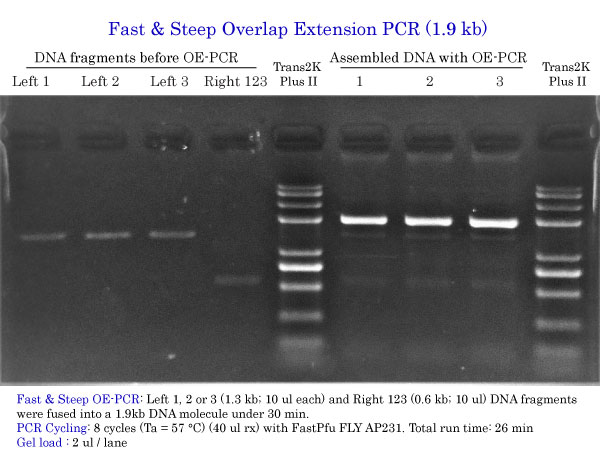
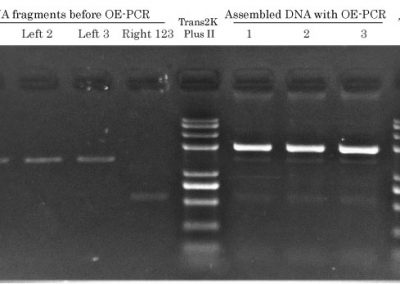
Left 1, 2 or 3 (1.3 kb) fragments, each amplified from their respective parent template containing only either one of the three mutations, and thee Right 123 (0.6 kb) DNA fragment, used to add a second and common mutation to any of the 1rst mutations, were amplified by Fast & Steep PCR.
PCR Setup :
ddH2O : to 35 ul
5x buffer: 7 ul
dNTPs (2,5 mM) : 2.8 ul
F primer (100 uM) : 0.14 ul (14 pmol)
R primer (100 uM) : 0.14 ul (14 pmol)
*Minicircle : 300 ng
FastPfu FLY : 0.7 ul (1.75 u)
PCR Cycling: 7 cycles (Ta = 62 °C)
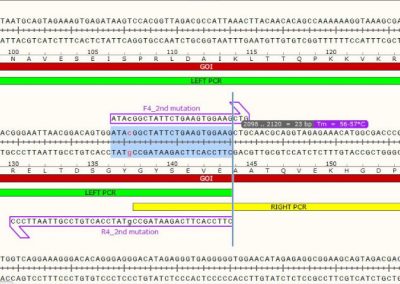
Overlap between Left and Right DNA fragments before OE-PCR
LEFT and RIGHT PCR fragments overlap each other by 23 bp. The overlapping region contains the second desired mutation in order to generate double mutants. The 23 bp overlap has an expected Tm of 57 °C. It’s important to design overlapping DNA segments having a Tm equal or greater than the Tm of external primers used for the OE-PCR reaction.
Fast & Steep OE-PCR 1.9 kb
PCR Setup :
ddH2O : to 40 ul
5x buffer: 8 ul
dNTPs (2,5 mM) : 3.2 ul
F primer (10 uM) : 1.6 ul (16 pmol)
R primer (10 uM) : 2 ul (16 pmol)
LEFT DNA : 10 ul
RIGHT DNA : 10 ul
FastPfu FLY : 0.8 ul (2 u)
PCR Cycling: 8 cycles (Ta = 57 °C) Total run time: 26 min Gel load : 2 ul / lane