Complete Design, Cloning and Truncation Analysis of a Human Promoter
PCR Success Story #15Project Description
Complete Design, Cloning and Truncation Analysis of a Human Promoter (paid service)
To protect the research and privacy of the customer who paid for our Expression Vector Preparation Service, the name of the gene promoter and researcher are held confidential.
This cloning project originated from a lead found while supporting the 31e Congrès de la Recherche – CHU Ste-Justine back in May 2016.
Our work consisted in
- promoter sequence analysis
- sequence truncation suggestions for promoter analysis by luciferase assay
- design of the cloning strategy
- performing the cloning of 12 segments (truncation method) into the TK-LUC expression vector provided by the researcher.
To perform the cloning experiments, we have used TransBionovo (formerly known as Transgen Biotech) products:
Project Details
Client: Centre de Recherche du CHU Ste-Justine
Date: August 22th, 2016
Type of experiment: Design and Cloning of a human promoter region
DNA Polymerase: TransStart FastPfu High-Fidelity DNA Polymerase and others
Promoter Truncation Analysis and Design
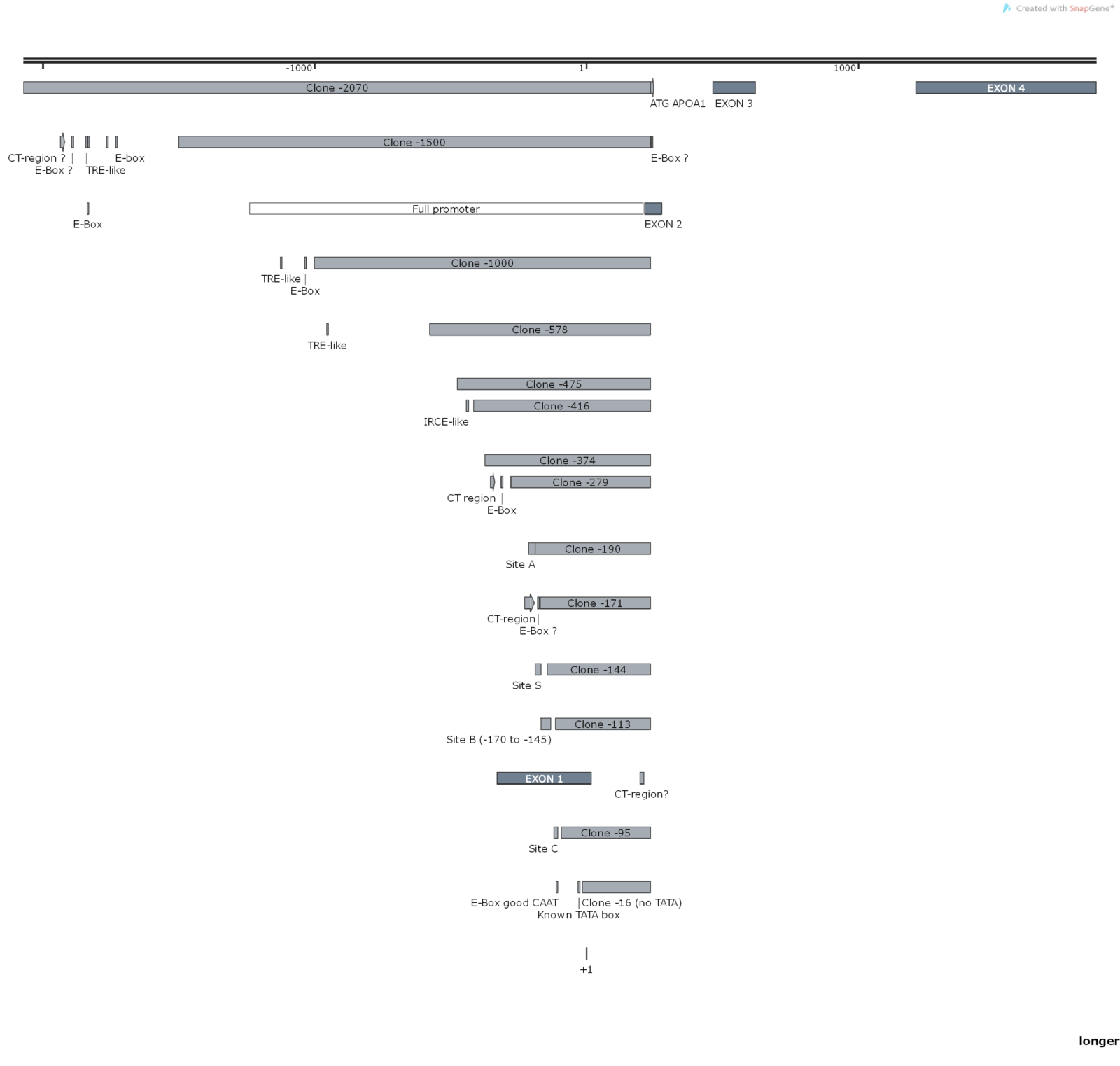
We’ve analyzed the sequence of the promoter region of the researcher’s gene of interest and have designed a truncation strategy that would enable him to find key regulatory regions within this promoter.
The researcher opted for 12 truncations of the full-lenght promoter
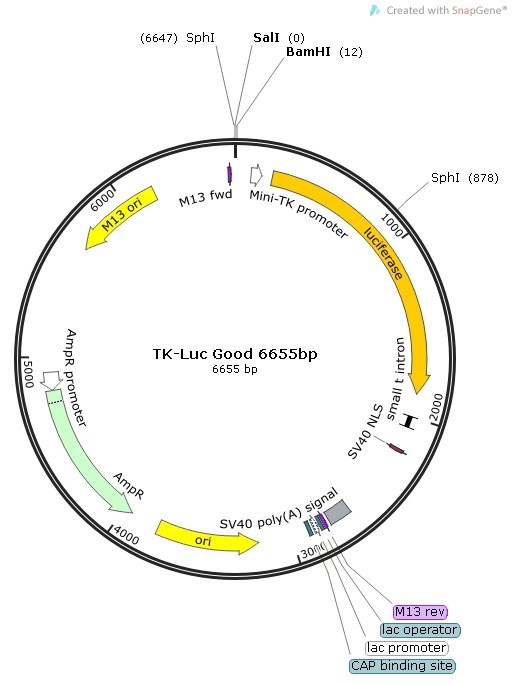
Design of the Cloning Strategy into TK-Luc Vector
Because none of the promoter truncation sequences contained either the restriction sites Sal I or Bam HI, we deisgned forward primers to contain a Sal I restriction site + 6 floating bases in its 5′ sequence (5′ of PCR products).
The reverse primer was universal for this cloning stategy and contained a Bam HI restriction + 6 floating bases in its 5′ sequence (3′ end of PCR fragments).
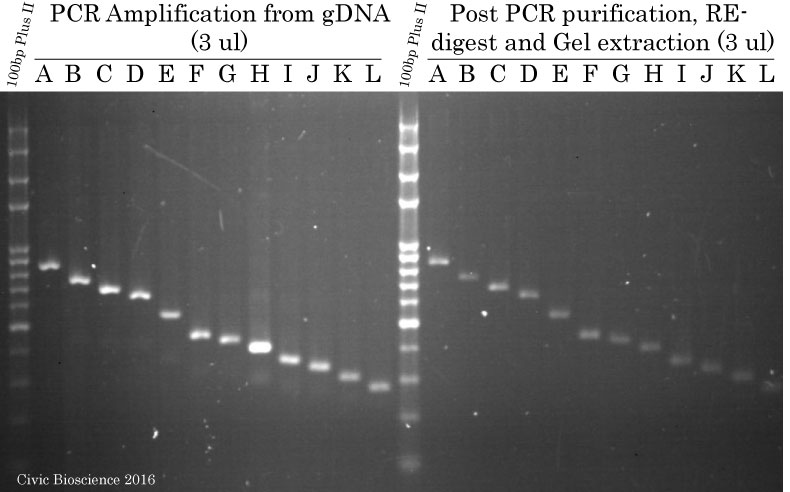
PCR Amplification of Promoter segments from gDNA using FastPfu DNA Polymerase
Promoter fragments were amplyfied by PCR using TransStart FastPfu (High-Fidelity) DNA Polymerase from purified human genomic DNA obtained from PCR Success Story #8. Subsequently, each amplicon was purified on column using the FavorPrep GEL/PCR Purification Kit, digested using FlyCut Sal I and Bam HI and finally gel extracted using the same FavorPrep GEL/PCR Purification Kit. Samples (3 ul) were run on a 1.6% agarose gel stained with ethidium bromide.
PCR reaction setup:
- H2O : to 50 ul
- 5x FastPfu Buffer 10 ul
- Forward primer (10 uM): 1 ul (200 nM final)
- Reverse primer (10 uM) : 1 ul (200 nM final)
- gDNA : 5 ul (50 ng)
- FastPfu: 1 ul (2.5 u)
PCR cycling (for all) :
- Denaturation: 60s at 95 °C
- 35 x
- Denaturation: 15s at 95 °C
- Annealing: 20s at 63 °C
- Extension: 15s at 72 °C
- Final extension: 120s at 72 °C
Ligation, Transformation and Restriction Analysis
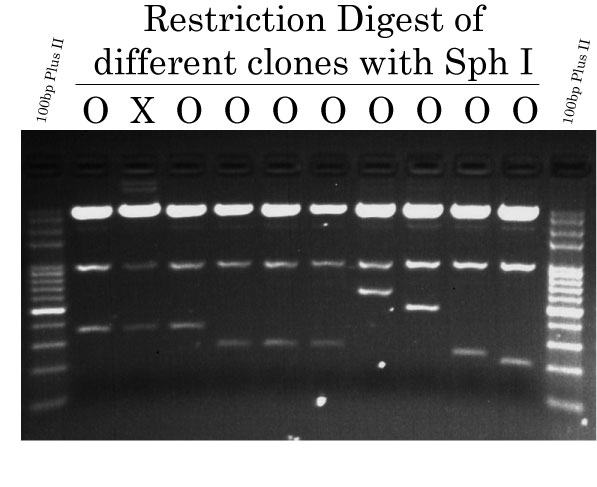
After also digesting the TK-Luc vector with Bam HI and Sal I (Bam HI-TK-Luc-Sal I), followed by gel extraction, the vector was ligated individually to each of the 12 Sal I-fragments-Bam HI using T4 DNA Ligase. Ligations were then transformed into Trans1-T1 Phage Chemically Competent Cells. The next day, colonies were screened by colony PCR using EasyTaq DNA Polymerase and then chosen colonies were picked up for plasmid miniprep DNA extractions. Restriction digest analysis was performed using FlyCut Sph I to determine wether the plasmid preparations were constructed as intended or not. Restriction digest were analyzed by agarose gel (1.8%) electrophoresis.
Sequencing
Plasmid miniprep were sent out for sequencing at La Plateforme de Séquençage du CHUL in Quebec City and then sent out to the researcher with all maps, sequences, minipreps, glycerol stocks.
Want us to Perform and Design your Cloning Project too?
Put us in charge !