Detection Limit of High-Fidelity DNA Polymerases on Human gDNA
High-Fidelity DNA Polymerase ComparisonDetection Limit of High-Fidelity DNA Polymerases using Human gDNA for Template
We previously determined the detection limit of different Taq DNA Polymerases using human genomic DNA as a template for PCR. For this instance, we aimed at determining the Detection Limit of High-Fidelity DNA Polymerases in similar conditions as described previously.
In the current assay, we performed multiplex PCR amplification or a human β-globin fragment (536 bp) and the full-lenght coding region of the Melanocortin-2 Receptor (MC2R; 891 bp) from purified human genomic DNA.
To determine the Detection Limit of High-Fidelity DNA Polymerases, the following masses of purified human genomic DNA were used: 25 ng, 2.5 ng, 250 pg or 25 pg.
KP*, KKP*, FastPfu and FastPfu FLY High-Fidelity DNA Polymerases and competitor DNA Polymerases: Phusion® HotStart II and Q5® High-Fidelity DNA Polymerases were compared.
Details
Date: January 29th, 2016
Type of experiment: Multiplex PCR from human gDNA
DNA Polymerase: KP*, KKP*, TransStart FastPfu and FastPfu FLY High-Fidelity DNA Polymerases
Competitor DNA Polymerases: Phusion® HotStart II and Q5® High-Fidelity DNA Polymerases
Phusion® is a registered trademark of ThermoFisher Scientific. Q5® is a registered trademark of New England Biolabs, Inc.
*KP and KKP are currently prototype versions of 2 new High-Fidelity DNA Polymerases that are under development.
Multiplex PCR setup for Detection Limit of High-Fidelity DNA Polymerases
- H2O : fill to 15 ul
- 5x buffer : 3 ul
- dNTPs (2.5 mM): 1.2 ul (0.2 mM final)
- β-globin Forward primer (10 uM): 0.3 ul (0.2 uM final)
- β-globin Reverse primer (10 uM) : 0.3 ul (0.2 uM final)
- MC2R Forward primer (10 uM): 0.3 ul (0.2 uM final)
- MC2R Reverse primer (10 uM) : 0.3 ul (0.2 uM final)
- gDNA : 1 ul (sequencially pre-diluted to 25000, 2500, 250 and 25 pg per ul)
- DNA Polymerase (2.5 u/ul) : 0.3 ul
Multiplex PCR setup for Detection Limit of High-Fidelity DNA Polymerases
- H2O : fill to 15 ul
- 5x buffer : 3 ul
- dNTPs (2.5 mM): 1.2 ul (0.2 mM final)
- β-globin Forward primer (10 uM): 0.75 ul (0.5 uM final)
- β-globin Reverse primer (10 uM) : 0.75 ul (0.5 uM final)
- MC2R Forward primer (10 uM): 0.75 ul (0.5 uM final)
- MC2R Reverse primer (10 uM) : 0.75 ul (0.5 uM final)
- gDNA : 1 ul (sequencially pre-diluted to 25000, 2500, 250 and 25 pg per ul)
- DNA Polymerase (2 u/ul) : 0.15 ul for Phusion and Q5; 0.3 ul for KP
Multiplex PCR cycling for establishing the Detection Limit of High-Fidelity DNA Polymerases
Denaturation: 60s at 95 °C
35 x
- Denaturation: 15s at 95 °C
- Annealing: 20s at 57 °C (KP, KKP, FastPfu, FLY) or 60 °C (Phusion, Q5)
- Extension: 25s at 72 °C
Final extension: 120s at 72 °C
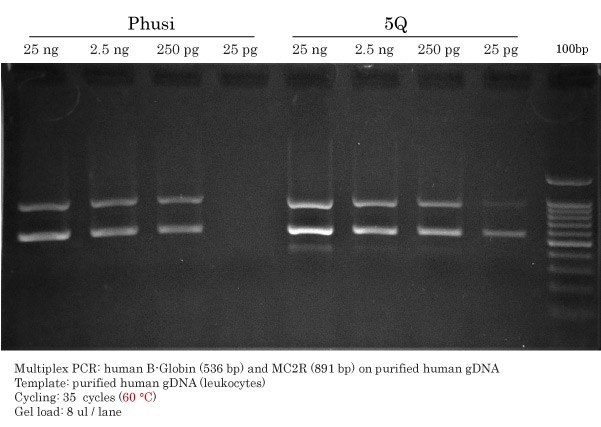
Phusion and Q5 Detection Limit on gDNA
First, the Detection Limit of High-Fidelity DNA Polymerase multiplex PCR was performed using competitor DNA Polymerases, namely Phusion® HotStart II and Q5® High-Fidelity DNA Polymerases. The annealing temperature used was 60 °C.
First, Phusion® was more specific than Q5®, although offering less DNA yield. However, Phusion® HotStart II and Q5® were able to amplify bands corresponding to human β-globin (536 bp) and MC2R (891 bp) using 25 ng, 2.5 ng and 250 pg of human genomic DNA. These DNA masses correspond to 7.5 x 103, 7.5 x 102 and 7.5 x 101 copies of target template respectively. Abruptly, Phusion® stopped ‘working’ with 25 pg gDNA, whereas Q5® detected β-globin. However, MC2R amplification using Q5 was very modest and unefficient using only 25 pg gDNA (7.5 copies).
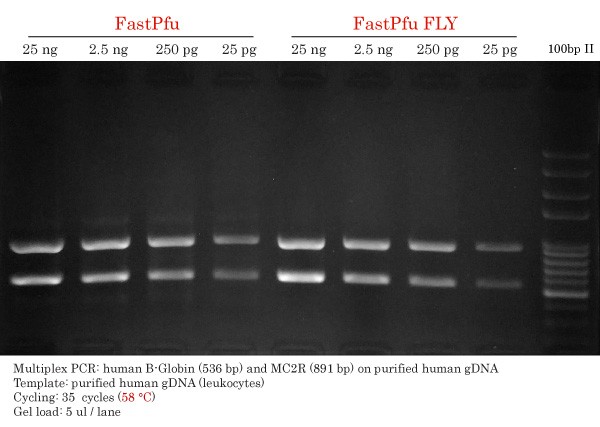
FastPfu and FastPfu FLY Detection Limit on gDNA
Secondly, Detection Limit of High-Fidelity DNA Polymerase multiplex PCR was performed using TransStart FastPfu and FastPfu FLY High-Fidelity DNA Polymerases. The annealing temperature used to obtain this result was 57 °C.
First, FastPfu gave similar DNA yield for MC2R amplification s compared FastPfu FLY using 57 °C as the annealing temperature. Nevertheless, FastPfu could detect human β-globin (536 bp) using all the gDNA input range.
Remarkably, FastPfu FLY was very specific in addition to successfully amplifying both β-globin (536 bp) and MC2R (891 bp). Each band (8 ul load) represently approximately 50 ng as compared to the 100bp DNA ladder. Simply superb performance (again)!
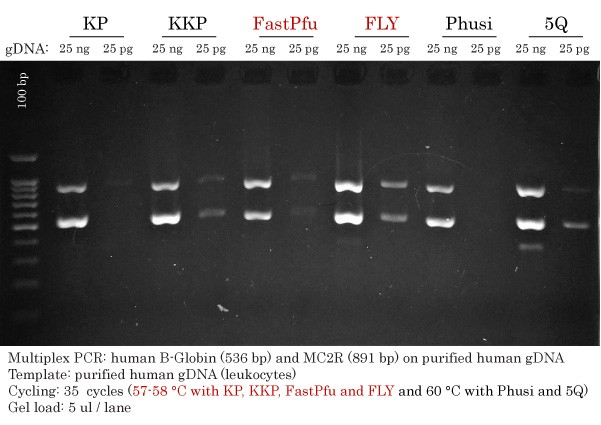
Summary of the Detection Limit of High-Fidelity DNA Polymerases
In one image, we recapitulated the results of the performance comparison and the Detection Limit of High-Fidelity DNA Polymerases on human gDNA. All amplicons obtained using either 25 ng or 25 pg with all tested High-Fidelity DNA Polymerase could then be compared side-by-side.
This comparison better illustrates the differences between each DNA Polymerase.
Ranking of High-Fidelity DNA Polymerases:
- Detection limit: FLY > Q5 = KKP > FastPfu > KP > Phusion
- Yield: FLY > Q5 ≥ KKP > KP ≥ FastPfu ≥ Phusion
Setting a Higher Level of DNA Polymerase Performance
Clearly, our results demonstrate that TransBionovo High-Fidelity DNA Polymerases set a Higher Level of DNA Polymerase Performance when compared to the popular products such as Phusion® and Q5® DNA Polymerases.
The WINNER of this ” Detection Limit of High-Fidelity DNA Polymerases ” comparison is: TransStart FastPfu FLY (Ultra-HiFi )DNA Polymerase.
In addition, KP and KKP DNA Polymerases, currently in development, have shown promising performance, albeit not being able to outperform the great FastPfu FLY!
Pick up the Rare DNA you are Looking for!

Is your HiFi good enough?
DNA Polymerase Comparison Chart
Specifications and pricing of commercially available Taq DNA Polymerases and High-Fidelity DNA Polymerases in Canada.
PCR Purification Kit Comparison – DNA Clean Up
Which is the Best Kit for DNA Clean Up? Our aim was to perform a PCR Purification Kit Comparison from different vendors. At Civic Bioscience, we distribute the Favorprep™ and EasyPure® product lines from FAVORGEN and TransBionovo respectively. First, the FavorPrep™...
miniPCR Review : How I use a portable PCR thermocycler to help life scientists day-to-day.
The miniPCR and blueGel systems represent the core instruments involved in the assistance we provide to real scientists who conduct life science research in Canada. Both laboratory instruments were essential in solving dozens of problems that prospect customers were...
GC-rich PCR: Comparison of FastPfu FLY and Q5 High-Fidelity DNA Polymerases
GC-rich PCR: FastPfu FLY vs Q5® High-Fidelity DNA Polymerase GC-rich PCR can be difficult to achieve Our aim was to verify compare FastPfu FLY (TransBionovo) and Q5 (NEB) high-fidelity DNA Polymerase at amplifying a GC-rich sequence (human ARX; 79% GC) from purified...
Detection Limit of Taq DNA Polymerase
What's the Detection Limit of Taq ? How many DNA copies from gDNA? There are MANY Taq DNA Polymerases out there on the market. Some are driven by strong marketing and others by strong performance. Our aim was to verify the Detection Limit of Taq DNA Polymerases in PCR...
Taq DNA Polymerase Comparison (Part 2)
Taq DNA Polymerase Comparison...which one should you choose? Nowadays, many suppliers offer Taq DNA Polymerases. Our aim was to perform a Taq DNA Polymerase Comparison. Our Taq DNA Polymerase Comparison assay consisted in PCR amplification of a fragment of the human...