Success #1: EGFP, flox/src, Cre, Confetti Mouse genotyping
PCR Success Story #1Project Description
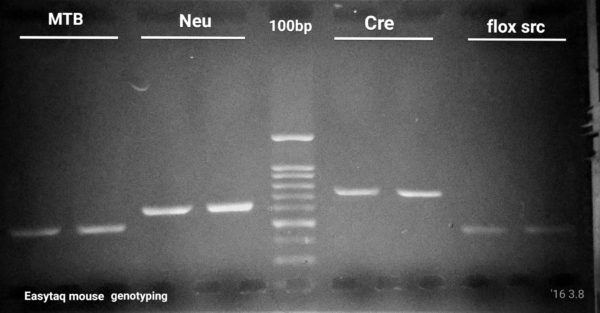
EGFP, flox/src, Cre, Confetti Genotyping by PCR
6 genes to amplify, 4 PCR cycling protocols and 1 missing band.
Our first challenge consisted in optimizing a genotyping PCR protocol for a well established lab located at McGill University. Over the years, the group had optimized PCR rections and cycling conditions for at least 6 genes. For these 6 genes, they had to use 2 different Taq DNA polymerases (NEB and Qiagen$$$) and 4 different PCR cycling protocols. Their group routinely performs EGFP, flox/src, NeuN, Cre,and Confetti Genotyping by PCR. However, their ”Confetti” PCR was the most challenging, often failing at amplifying the mutant allele, even with the most expensive Taq DNA polymerase on the market (supplier Q). Cost was an issue, but performance was critical. The researcher provided us with PCR primers and gDNA samples for 6 routine genotyping testing including their hard-to-get ”Confetti”. Please find bellow a resume of what we have accomplished using Transgen Biotech’s EasyTaq® DNA Polymerase.
Project Details
Client: McGill University
Date: March 8th, 2016
Type of experiment: Mouse genotyping by PCR
DNA Polymerase: EasyTaq DNA Polymerase.
Competitors: NEB and Qiagen Taq DNA polymerases
EGFP, flox/src, Cre, NeuN Genotyping by PCR
5 / 6 perfect PCR reactions in 1 common PCR setup and cycling protocol
Even if primer pair melting temperatures (Tm) were 5-6 °C apart, we managed to perfectly optimize 5 out of 6 (4 shown here) PCR reactions using the exact same cycling protocol. In addition, we’re shortened the PCR run time by ∼30 min as compared to the PCR cycling protocol the researcher was using. This allowed the researcher to save time on their routine genotyping experiments.
Confetti Genotyping by PCR
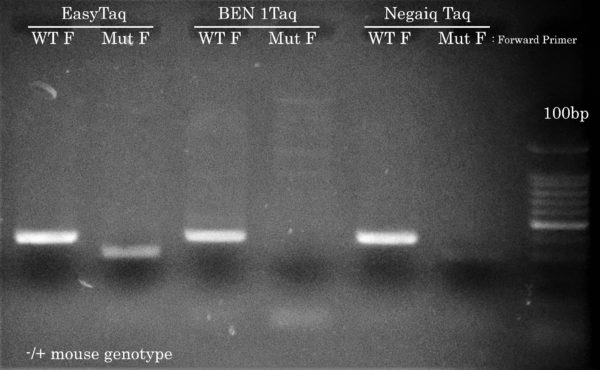
Only EasyTaq can amplify the mutant Confetti allele
Using the same PCR cycling protocol as the other 5 genotyping PCR reactions but using a lower Ta of 54°C and using either a ”WT” or ”mutant” forward primer, EasyTaq, but not Taq DNA polymerases from competitors, achieved Confetti Genotyping by PCR of the mutant allele from the hemizygous mouse gDNA sample. This result is crucial for the McGill researcher in order to differentiate the different mice genotypes used in their lab. All Taq DNA polymerases correctly amplified the WT allele.
Confetti PCR setup
- H2O: to 20ul
- 10x : 2ul
- dNTPs (2.5mM): 1.6 ul
- F primer (10uM): 0.4ul (200nM final)
- R primer (10uM): 0.4ul
- gDNA: 0.5 to 2ul (1ul with my extracts)
- Easytaq: 0.2 ul (1u)
Confetti PCR Cycling
- 94 for 60s
35x
- 94 for 15s
- 54 for 20s
- 72 for 10s
Final extension 72 for 120s.
Genotyping by PCRGenotyping is easier with EasyTaq
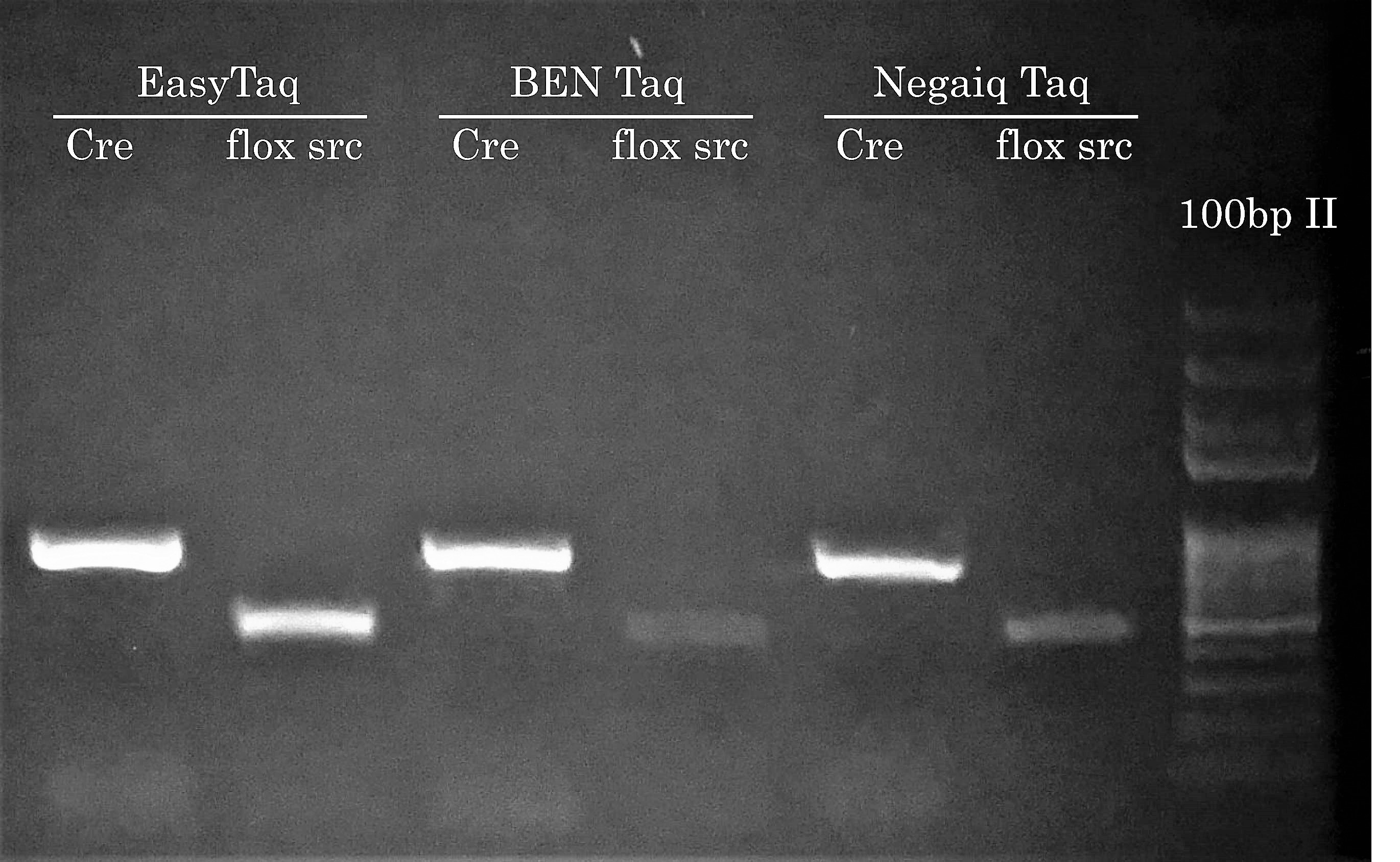
We know what you’re thinking: Hey Civic Bio, show us how competitor Taqs performed on easier PCR reactions. Well, we actually think about this during every project because performing PCR reactions with EasyTaq (or other Transgen Biotech DNA polymerase) feels almost like EasyTaq should be named ‘Magic Taq’ (we’ve actually stolen this expression from a customer – in PCR Success Story #2). Most of the time during a project, comparing competitor DNA polymerases is somewhat out of scope because the main goal is to solve problematic PCR reactions for our customers. Plus, seing competitor DNA polymerases not doing so great has become boring if you ask us… Who wants to see PCR reactions fail?! Not us.
Anyhow, this time we’ve performed a comparison with BEN Taq (∼ 80$ for 400u) and Neqaiq Taq (∼ 520$ for 1000u) DNA Polymerases. The genotype of these mice are positive for the indicated PCR targets. We’ll let you choose the DNA polymerase you feel should make your life easier!