High-Fidelity PCR Amplification of Pseudomonas aeruginosa murC and murE
PCR Success Story #14Project Description
High-Fidelity PCR Amplification of Pseudomonas aeruginosa murC and murE
This cloning project originated from the lab of a new researcher located at the main campus of Université de Montréal. It consisted in optimization of PCR conditions for the amplification of murC and murE from Pseudomonas aeruginosa. Cloning primers were designed by the customer in order to perform High-Fidelity PCR Amplification from Pseudomonas gDNA.
Pseudomonas aeruginosa genomic DNA was extracted from 1 colony with TransDirect Animal Tissue AD1, AD2, AD3 buffer set but was not purified.
Primer sequences were as follows. Letters in bold represent the gene-specific parts of each primer; GC% and Tm are indicated for the gene-specific regions:3
murC (1439 bp)
- Forward: tgacaagctttcagtgatggtgatggtgatggtgatggtgatgagagccaccgccacctgcgcccttccctcc (annealed bases: 73% GC; Tm = 57 °C)
- Reverse: gtcacatatggttaaagaaccgaatggcgtca (annealed bases: 48% GC; Tm = 60.5 °C)
murE (1461 bp)
- Forward: tgacaagctttcagtgatggtgatggtgatggtgatggtgatgagagagccaccgccaccagcatgcggcacctcc (annealed bases: 69% GC; Tm = 60.5 °C)
- Reverse: gtcacatatgcctatgagcctgaaccaactg (annealed bases: 50% GC; Tm = 60.5 °C)
Project Details
Client: Université de Montréal
Date: June 10th, 2016
Type of experiment: High-Fidelity amplification from Pseudomonas aeruginosa gDNA
DNA Polymerases: TransStart FastPfu and FastPfu FLY
Competitors: None
PCR Amplification of murC using FastPfu
The murC gene has an overall GC content of 67% GC with some segments reaching 75% GC.
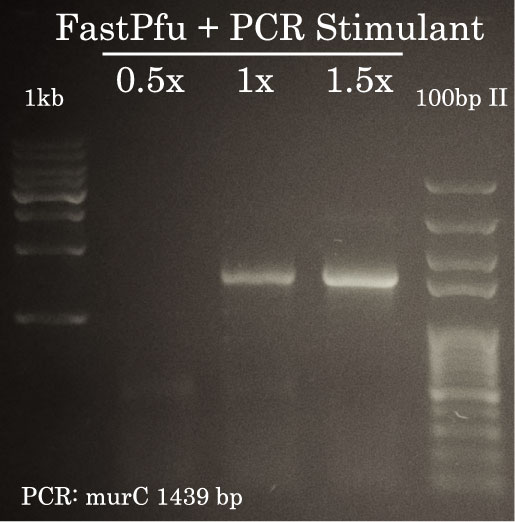
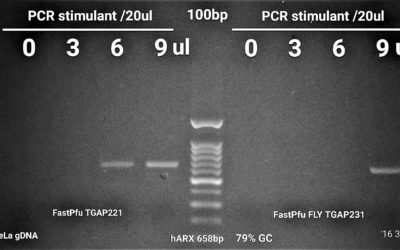
TransStart FastPfu was used to amplify murC from Pseudomonas aeruginosa genomic DNA. As illustrated here, successful amplification necessitated the addition of PCR Stimulant. 1.5 ul of the PCR reaction was migrated on a 1% agarose gel in TAE 1x.
PCR setup for High-Fidelity PCR:
- H2O : to 50 ul
- 5x buffer : 10 ul
- PCR Stimulant (5x): 5, 10 or 15 ul
- dNTPs (2.5mM): 4 ul (0.2 mM final)
- F1 (10 uM): 2 ul (200 nM final)
- F2 (10 uM): 2 ul
- cDNA : 5 ul of P.aeruginosa colony gDNA extrcted with TransDirect Animal Tissue Buffer Set
- FastPfu (2.5 u/ul) : 1 ul
PCR cycling for High-Fidelity PCR
- Denaturation: 60s at 98 °C
- 35 x
- Denaturation: 15s at 98 °C
- Annealing: 35s at 54 °C
- Extension: 40s at 72 °C
- Final extension: 300s at 72 °C
High-Fidelity PCR Amplification of murE using FastPfu and FastPfu FLY
The murE gene has an overall GC content of 69% GC with some segments reaching 80% GC.
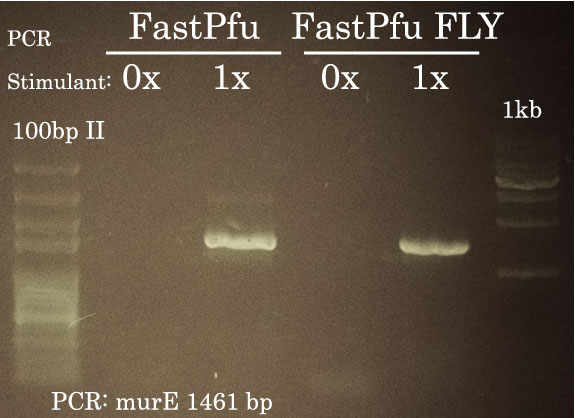
TransStart FastPfu was used to perform High-Fidelity PCR and amplify murE from Pseudomonas aeruginosa genomic DNA. As illustrated here, successful amplification necessitated the addition of PCR Stimulant to 1x fnal concentration. 1ul of the PCR reaction was migrated on a 1% agarose gel in TAE 1x.
PCR reaction setup:
- H2O : to 50 ul
- 5x buffer : 10 ul
- PCR Stimulant (5x): 10 ul
- dNTPs (2.5mM): 4 ul (0.2 mM final)
- F1 (10 uM): 2 ul (200 nM final)
- F2 (10 uM): 2 ul
- cDNA : 5 ul of P.aeruginosa colony gDNA extrcted with TransDirect Animal Tissue Buffer Set
- FastPfu (2.5 u/ul) : 1 ul
PCR cycling :
- Denaturation: 60s at 98 °C
- 35 x
- Denaturation: 15s at 98 °C
- Annealing: 35s at 52 °C
- Extension: 40s at 72 °C
- Final extension: 300s at 72 °C
Problems with high-fidelity PCR?
High-Fidelity DNA Polymerase Comparison using unpurified gDNA
High-Fidelity DNA Polymerase Comparison on difficult genomic DNA samples. Modern commercially available high-fidelity DNA Polymerases are derived from either Pfu and KOD. Pfu is an enzyme found in the hyperthermophilic archaeon Pyrococcus furiosus. KOD is an enzyme...
Direct PCR on hair, saliva, cheek scrub and dead skin lysates
TransDirect® Animal Tissue Direct PCR Kit Genotyping is the process of identifying differences in the genetic make-up (genotype) of an individual by examining the individual's DNA sequence and comparing it to another specimen or a reference sequence. It reveals the...
Amplification of 79% GC-rich human ARX
Introduction ARX is expressed in a wide variety of tissues including the brain, heart, skeletal muscle, testis, intestine, and pancreas [1–2]. The human ARX gene has five exons that together encode a number of protein domains of the transcription factor. These include...
‘FLY’ your PCR | DNA Polymerase Extension Rate – 6kb/min
Simply 'FLY' your long PCRs - Polymerase Extension Rate Transgen Biotech manufactures three High-Fidelity DNA Polymerases, TransStart® KD Plus, FastPfu and FastPfu FLY. TransStart® FastPfu FLY, a ultra-high fidelity (108x higher fidelity than EasyTaq) and ultra-high...