Base Deletion by Site-Directed Mutagenesis with Extreme GC Content
PCR Success Story #12Project Description
Small 1 base deletion from an Extreme GC Content template (up to 94% GC-rich)
This project originated from previous exchanges with a customer having problems with Extreme GC Content PCR amplification. Their plasmid codes for a sfGFP-GOI in pcDNA3. The researcher’s lab is located at the the Université de Montréal in Montreal. The problem is that their GOI has Extreme GC Content. The first 200 bp of hHCN2 has a minimal GC content of 92%.Then, the GC content does not drop below 76% for at least another 400 bp. Overall, the 3006 bp coding for the GOI has a GC content of 70%. Total vector size is 9108 bp. That sure sounded like a nice challenge for us to solve!
Project Details
Client: Université de Montréal
Date: June 5th, 2016
Type of experiment: 1 base deletion by Site-Directed Mutagenesis in an extremely GC-rich gene
DNA Polymerase: TransStart FastPfu High-Fidelity DNA Polymerase &
Competitor: None
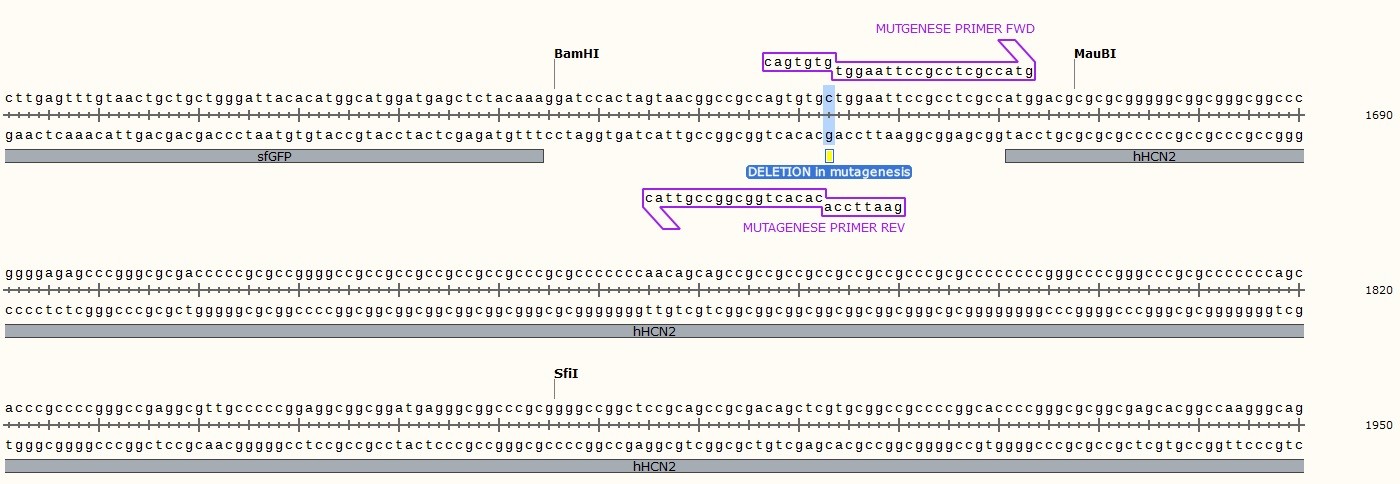
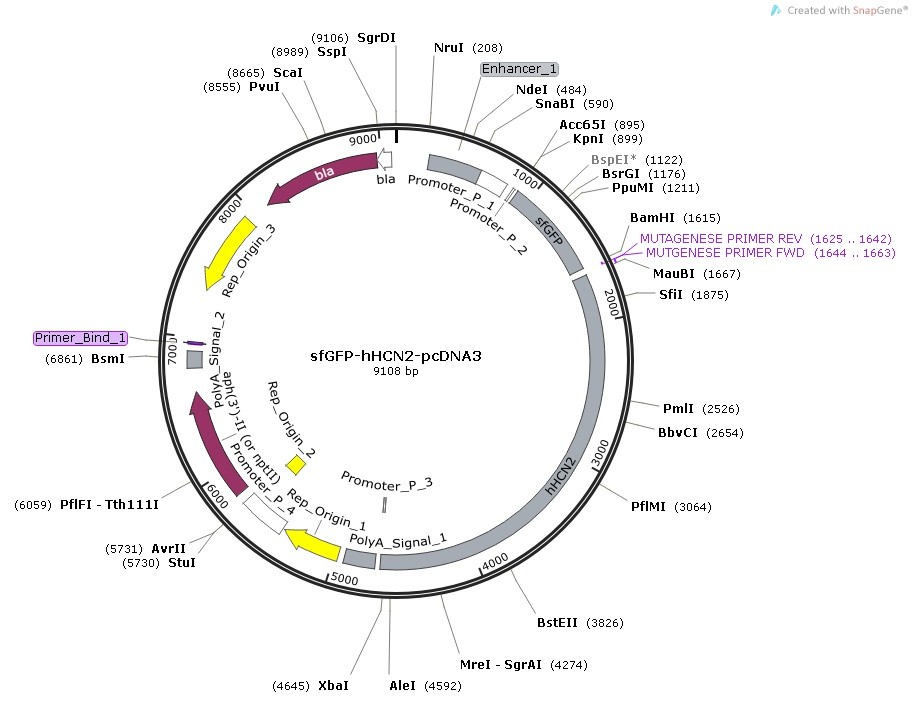
Plasmid Map and sequence of sfGFP-hHCN2/pcDNA3
sfGFP-hHCN2 in pcDNA3 (9108 bp)
The first 200 bp of hHCN2 has a minimal GC content of 92%.Then, the GC content does not drop below 76% for at least another 425 bp. Overall, the 3006 bp coding for hHCN2 has a GC content of 70%.
Primers:
F: cagtgtgtggaattccgcctcgccatg
R: gaattccacacactggcggccgttac
High GC content isn't difficult when you can FLY!
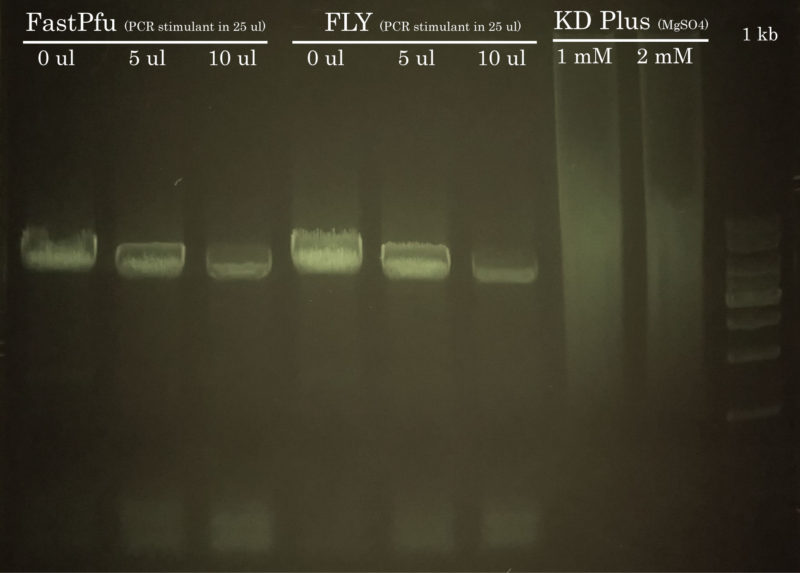
Way too much DNA was loaded on the gel, but hey! we got it (again) on our first attempt.
The mutagenesis PCR was performed using either TransStart FastPfu or FastPfu FLY in presence or not of increasing amounts of PCR Stimulant. PCR using KD Plus also was attempted but you can see that it failed. The presence of PCR stimulant only lowered the yield of the reaction, therefore it was not necessary to use it.
Mutagenesis PCR reaction setup:
- H2O : complete to 25 ul
- 5x buffer: 5 ul
- dNTPs (2.5 mM): 2 ul
- Forward primer (10 uM): 0.5 ul (200 nM final)
- Reverse primer (10 uM) : 0.5 ul (200 nM final)
- sfGFP-hHCN2/pcDNA3 plasmid : 1 ul (10 ng)
- PCR Stimulant: 0, 5 or 10 ul
- Polymerase: 0.5 ul
PCR cycling for site-directed mutagenesis :
- Denaturation: 120s at 94 °C
- 25 x
- Denaturation: 30s at 94 °C
- Annealing: 20s at 58 °C
- Extension: 500s at 72 °C
- Final extension: 600s at 72 °C
Available Products for Site-Directed Mutagenesis
-
DMT Competent Cell (DpnI+) – CD511
$136.00 – $212.00 CAD -
DMT Enzyme ( Dpn I ) – GD111
$72.00 CAD -
Fast MultiSite Mutagenesis System – FM201
$600.00 CAD -
Fast Mutagenesis System – FM111
$240.00 – $415.00 CAD -
Ju™ DNA Assembly Mix for Cloning and Mutagenesis – CB-JU101
$125.00 – $280.00 CAD - Sale!
KLD Mix for Back-to-Back Site-Directed Mutagenesis
$110.00 – $464.00 CAD
Want PCR to be easier for you too?
Put us in charge of optimizing PCR conditions for you. It’s FREE !