KLD Site-Directed Mutagenesis Protocol
Site-Directed Mutagenesis using WVA, back-to-back primers and KLD Build your own KLD Mutagenesis KitGo to the Main SDM PageKLD Site-Directed Mutagenesis Protocol using Back-to-Back Primers
Advantages of KLD Site-Directed Mutagenesis
- Simple to use KLD mix
- High-yield exponential PCR.
- Simple primer design.
- In vitro digestion of the template DNA.
- 90% of clones contain the desired mutation.
- Suitable for insertions of ∼ 80 bases or longer if using oligos > 60-mers.
- Ideal for creating large deletions.
- Easy 3-step workflow***.
Disadvantages
- Long PCR protocol – 25 cycles (between 4 and 8 hours or 1 to 2 hours using Fast & Steep PCR).
- Only 1 primer contains the mutation which may generate non-methylated and non-mutated PCR products.
- Primers or Dpn I-generated fragments are likely to be inserted at the ligation site.
***To ensure that no extra mutations are introduced in the vector backbone, site-directed mutagenesis strategies, such as the herein KLD strategy, which use whole vector amplification require subcloning of the sequenced insert into a vector that hasn’t been amplified by PCR. Alternatively, the whole vector can be sequenced.
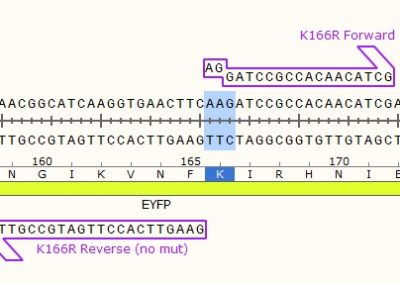
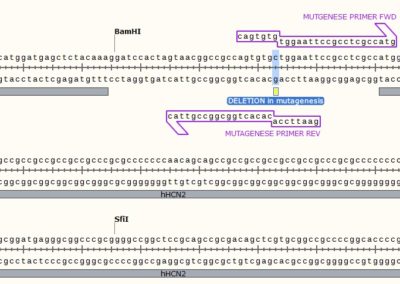
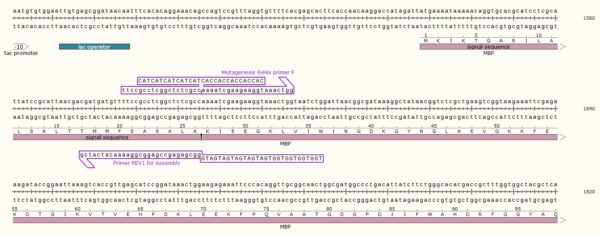
STEP 1 - Primer Design
Point Mutation
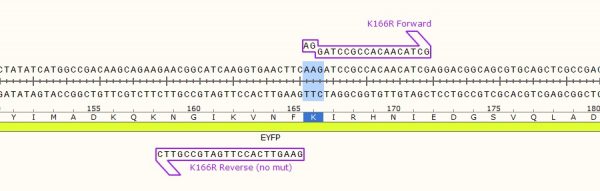
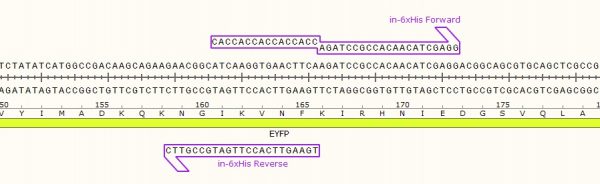
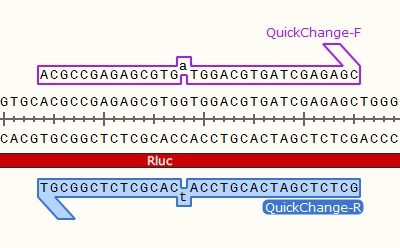
As in the name of the technique and in contrast to QuickChange and Fast Mutagenesis primer design, both primers must be designed ‘back-to-back’ in opposite directions. Only one (1) of the primers must contain the desired point mutation.
STEP 2 - PCR
WVA PCR Protocol for KLD Mutagenesis
Classic WVA PCR Setup
- H2O : to 25 ul
- 5x buffer : 5 ul
- dNTPs (2.5 mM): 2 ul (0.2 mM final)
- Forward Primer (10 uM): 0.5 ul (200 nM final)
- Reverse Primer (10 uM): 0.5 ul (200 nM final)
- plasmid DNA : 1-10 ng
- FastPfu FLY (2.5 u/ul) : 0.5 ul
PCR Cycling for WVA
- Denaturation: 120s at 95 °C
- 16-25 x
- Denaturation: 20s at 95 °C
- Annealing: 20s at Tm °C
- Extension: 10-30 s/kb at 68 °C
- Final extension: 600s at 68 °C
Want to go Faster and Save Time?
Use the Fast & Steep PCR protocol for whole vector amplification (WVA) to limit the number of PCR cycles to a minimum and achieve high-yield PCR. The risk of introducing undesired mutations drops drastically by at least 100 000-fold.
STEP 3 - KLD Treatment
KLD : Removal of Methylated Template, Phosphorylation and Ligation
Mix 6 ul sterile ddH2O Save & Exit
+ 1 ul of PCR product
+ 2 ul of 5x KLD buffer
+ 1 ul of KLD mix at RT for 5-15 min.
Incubate at room temperature for 5-15 min.
STEP 4 - Transformation
Transformation of Competent Cells
- Thaw 50 ul of high-efficiency (> 108 cfu/ug) chemically competent cells on ice.
- Add 1-2 ul of the KLD reaction from Step 3 and gently flick the tube 3 times before incubating on ice for 30 min.
- Heat shock the cells by at precisely 42 °C for 30-45 s (depends on the cells).
- Incubate on ice for > 2 min.
- Add 450 ul of SOC or LB media to the cells, then agitate at 200 rpm, 37 °C for 45-60 min.
- Spread 50-200 ul on a prewarmed LB-agar plate containing the appropriate antibiotic(s).
Optimal - Transformation in DMT Competent Cells
Add 1-5 μl of the KLD reaction to 50 μl of DMT chemically competent cells (CD511).
• Incubate on ice for 30 minutes.
• Heat shock at 42°C for 45 seconds.
• Incubate on ice for 2 minutes.
• Add 450 μl SOC or LB, gently shake at 37°C for 1 hour.
Spread 20–200 μl onto appropriate selection plate, incubate overnight at 37°C